Archive
Chinchilla Scaling: A replication using the pdf
The paper Chinchilla Scaling: A replication attempt by Besiroglu, Erdil, Barnett, and You caught my attention. Not only a replication, but on the first page there is the enticing heading of section 2, “Extracting data from Hoffmann et al.’s Figure 4”. Long time readers will know of my interest in extracting data from pdfs and images.
This replication found errors in the original analysis, and I, in turn, found errors in the replication’s data extraction.
Besiroglu et al extracted data from a plot by first converting the pdf to Scalable Vector Graphic (SVG) format, and then processing the SVG file. A quick look at their python code suggested that the process was simpler than extracting directly from an uncompressed pdf file.
Accessing the data in the plot is only possible because the original image was created as a pdf, which contains information on the coordinates of all elements within the plot, not as a png or jpeg (which contain information about the colors appearing at each point in the image).
I experimented with this pdf-> svg -> csv route and quickly concluded that Besiroglu et al got lucky. The output from tools used to read-pdf/write-svg appears visually the same, however, internally the structure of the svg tags is different from the structure of the original pdf. I found that the original pdf was usually easier to process on a line by line basis. Besiroglu et al were lucky in that the svg they generated was easy to process. I suspect that the authors did not realize that pdf files need to be decompressed for the internal operations to be visible in an editor.
I decided to replicate the data extraction process using the original pdf as my source, not an extracted svg image. The original plots are below, and I extracted Model size/Training size for each of the points in the left plot (code+data):
What makes this replication and data interesting?
Chinchilla is a family of large language models, and this paper aimed to replicate an experimental study of the optimal model size and number of tokens for training a transformer language model within a specified compute budget. Given the many millions of £/$ being spent on training models, there is a lot of interest in being able to estimate the optimal training regimes.
The loss model fitted by Besiroglu et al, to the data they extracted, was a little different from the model fitted in the original paper:
Original: 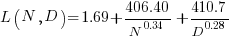
Replication: 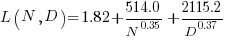
where: 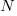 is the number of model parameters, and
is the number of model parameters, and 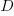 is the number of training tokens.
is the number of training tokens.
If data extracted from the pdf is different in some way, then the replication model will need to be refitted.
The internal pdf operations specify the x/y coordinates of each colored circle within a defined rectangle. For this plot, the bottom left/top right coordinates of the rectangle are: (83.85625, 72.565625), (421.1918175642, 340.96202) respectively, as specified in the first line of the extracted pdf operations below. The three values before each rg operation specify the RGB color used to fill the circle (for some reason duplicated by the plotting tool), and on the next line the /P0 Do is essentially a function call to operations specified elsewhere (it draws a circle), the six function parameters precede the call, with the last two being the x/y coordinates (e.g., x=154.0359138125, y=299.7658568695), and on subsequent calls the x/y values are relative to the current circle coordinates (e.g., x=-2.4321790463 y=-34.8834544196).
Q Q q 83.85625 72.565625 421.1918175642 340.96202 re W n 0.98137749 0.92061729 0.86536915 rg 0 G 0.98137749 0.92061729 0.86536915 rg 1 0 0 1 154.0359138125 299.7658568695 cm /P0 Do 0.97071849 0.82151775 0.71987163 rg 0.97071849 0.82151775 0.71987163 rg 1 0 0 1 -2.4321790463 -34.8834544196 cm /P0 Do |
The internal pdf x/y values need to be mapped to the values appearing on the visible plot’s x/y axis. The values listed along a plot axis are usually accompanied by tick marks, and the pdf operation to draw these tick marks will contain x/y values that can be used to map internal pdf coordinates to visible plot coordinates.
This plot does not have axis tick marks. However, vertical dashed lines appear at known Training FLOP values, so their internal x/y values can be used to map to the visible x-axis. On the y-axis, there is a dashed line at the 40B size point and the plot cuts off at the 100B size (I assumed this, since they both intersect the label text in the middle); a mapping to the visible y-axis just needs two known internal axis positions.
Extracting the internal x/y coordinates, mapping them to the visible axis values, and comparing them against the Besiroglu et al values, finds that the x-axis values agreed to within five decimal places (the conversion tool they used rounded the 10-digit decimal places present in the pdf), while the y-axis values appeared to differ differed by about 10%.
I initially assumed that the difference was due to a mistake by me; the internal pdf values were so obviously correct that there had to be a simple incorrect assumption I made at some point. Eventually, an internal consistency check on constants appearing in Besiroglu et al’s svg->csv code found the mistake. Besiroglu et al calculate the internal y coordinate of some of the labels on the y-axis by, I assume, taking the internal svg value for the bottom left position of the text and adding an amount they estimated to be half the character height. The python code is:
y_tick_svg_coords = [26.872, 66.113, 124.290, 221.707, 319.125] y_tick_data_coords = [100e9, 40e9, 10e9, 1e9, 100e6] |
The internal pdf values I calculated are consistent with the internal svg values 26.872, and 66.113, corresponding to visible y-axis values 100B and 40B. I could not find an accurate means of calculating character heights, and it turns out that Besiroglu et al’s calculation was not accurate.
I published the original version of this article, and contacted the first two authors of the paper (Besiroglu and Erdil). A few days later, Besiroglu replied with details of why they thought that the 40B line I was using as a reference point was actually at either 39.5B or 39.6B (based on published values for the Gopher budget on the x-axis), but there was uncertainty.
What other information was available to resolve the uncertainty? Ah, the right plot has Model size on the x-axis and includes lines that appear to correspond with axis values. The minimum/maximum Model size values extracted from the right plot closely match those in the original paper, i.e., that ’40B’ line is actually at 39.554B (mapping this difference from a log scale is enough to create the 10% difference in the results I calculated).
My thanks to Tamay Besiroglu and Ege Erdil for taking the time to explain their rationale.
The y-axis uses a log scale, and the ratio of the distance between the 10B/100B virtual tick marks and the 40B/100B virtual tick marks should be 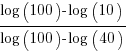 . The Besiroglu et al values are not consistent with this ratio; consistent values below (code+data):
. The Besiroglu et al values are not consistent with this ratio; consistent values below (code+data):
# y_tick_svg_coords = [26.872, 66.113, 124.290, 221.707, 319.125] y_tick_svg_coords = [26.872, 66.113, 125.4823, 224.0927, 322.703] |
When these new values are used in the python svg extraction code, the calculated y-axis values agree with my calculated y-axis values.
What is the equation fitted using these corrected Model size value? Answer below:
Replication: 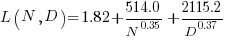
Corrected size: 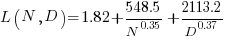
The replication paper also fitted the data using a bootstrap technique. The replication values (Table 1), and the corrected values are below (standard errors in brackets; code+data):
Parameter Replication Corrected
A 482.01 370.16
(124.58) (148.31)
B 2085.43 2398.85
(1293.23) (1151.75)
E 1.82 1.80
(0.03) (0.03)
α 0.35 0.33
(0.02) (0.02)
β 0.37 0.37
(0.02) (0.02) |
where the fitted equation is: 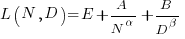
What next?
The data contains 245 rows, which is a small sample. As always, more data would be good.
A study, a replication, and a rebuttal; SE research is starting to become serious
tldr; A paper makes various claims based on suspect data. A replication finds serious problems with the data extraction and analysis. A rebuttal paper spins the replication issues as being nothing serious, and actually validating the original results, i.e., the rebuttal is all smoke and mirrors.
When I first saw the paper: A Large-Scale Study of Programming Languages and Code Quality in Github, the pdf almost got deleted as soon as I started scanning the paper; it uses number of reported defects as a proxy for code quality. The number of reported defects in a program depends on the number of people using the program, more users will generate more defect reports. Unfortunately data on the number of people using a program is extremely hard to come by (I only know of one study that tried to estimate number of users); studies of Java have also found that around 40% of reported faults are requests for enhancement. Most fault report data is useless for the model building purposes to which it is put.
Two things caught my eye, and I did not delete the pdf. The authors have done good work in the past, and they were using a zero-truncated negative binomial distribution; I thought I was the only person using zero-truncated negative binomial distributions to analyze software engineering data. My data analysis alter-ego was intrigued.
Spending a bit more time on the paper confirmed my original view, it’s conclusions were not believable. The authors had done a lot of work, this was no paper written over a long weekend, but lots of silly mistakes had been made.
Lots of nonsense software engineering papers get published, nothing to write home about. Everybody gets writes a nonsense paper at some point in their career, hopefully they get caught by reviewers and are not published (the statistical analysis in this paper was probably above the level familiar to most software engineering reviewers). So, move along.
At the start of this year, the paper: On the Impact of Programming Languages on Code Quality: A Reproduction Study appeared, published in TOPLAS (the first was in CACM, both journals of the ACM).
This replication paper gave a detailed analysis of the mistakes in data extraction, and the sloppy data analyse performed in the original work. Large chunks of the first study were cut to pieces (finding many more issues than I did, but not pointing out the missing usage data). Reading this paper now, in more detail, I found it a careful, well argued, solid piece of work.
This publication is an interesting event. Replications are rare in software engineering, and this is the first time I have seen a take-down (of an empirical paper) like this published in a major journal. Ok, there have been previous published disagreements, but this is machine learning nonsense.
The Papers We Love meetup group ran a mini-workshop over the summer, and Jan Vitek gave a talk on the replication work (unfortunately a problem with the AV system means the videos are not available on the Papers We Love YouTube channel). I asked Jan why they had gone to so much trouble writing up a replication, when they had plenty of other nonsense papers to choose from. His reasoning was that the conclusions from the original work were starting to be widely cited, i.e., new, incorrect, community-wide beliefs were being created. The finding from the original paper, that has been catching on, is that programs written in some languages are more/less likely to contain defects than programs written in other languages. What I think is actually being measured is number of users of the programs written in particular languages (a factor not present in the data).
Yesterday, the paper Rebuttal to Berger et al., TOPLAS 2019 appeared, along with a Medium post by two of the original authors.
The sequence: publication, replication, rebuttal is how science is supposed to work. Scientists disagree about published work and it all gets thrashed out in a series of published papers. I’m pleased to see this is starting to happen in software engineering, it shows that researchers care and are willing to spend time analyzing each others work (rather than publishing another paper on the latest trendy topic).
From time to time I had considered writing a post about the first two articles, but an independent analysis of the data meant some serious thinking, and I was not that keen (since I did not think the data went anywhere interesting).
In the academic world, reputation and citations are the currency. When one set of academics publishes a list of mistakes, errors, oversights, blunders, etc in the published work of another set of academics, both reputation and citations are on the line.
I have not read many academic rebuttals, but one recurring pattern has been a pointed literary style. The style of this Rebuttal paper is somewhat breezy and cheerful (the odd pointed phrase pops out every now and again), attempting to wave off what the authors call general agreement with some minor differences. I have had some trouble understanding how the rebuttal points discussed are related to the problems highlighted in the replication paper. The tone of the medium post is that there is nothing to see here, let’s all move on and be friends.
An academic’s work is judged by the number of citations it has received. Citations are used to help decide whether someone should be promoted, or awarded a grant. As I write this post, Google Scholar listed 234 citations to the original paper (which is a lot, most papers have one or none). The abstract of the Rebuttal paper ends with “…and our paper is eminently citable.”
The claimed “Point-by-Point Rebuttal” takes the form of nine alleged claims made by the replication authors. In four cases the Claim paragraph ends with: “Hence the results may be wrong!”, in two cases with: “Hence, FSE14 and CACM17 can’t be right.” (these are references to the original conference and journal papers, respectively), and once with: “Thus, other problems may exist!”
The rebuttal points have a tenuous connection to the major issues raised by the replication paper, and many of them are trivial issues (compared to the real issues raised).
Summary bullet points (six of them) at the start of the Rebuttal discuss issues not covered by the rebuttal points. My favourite is the objection bullet point claiming a preference, in the replication, for the use of the Bonferroni correction rather than FDR (False Discovery Rate). The original analysis failed to use either technique, when it should have used one or the other, a serious oversight; the replication is careful and does the analysis using both.
I would be very surprised if the Rebuttal paper, in its current form, gets published in any serious journal; it’s currently on a preprint server. It is not a serious piece of work.
Somebody who has only read the Rebuttal paper would take away a strong impression that the criticisms in the replication paper were trivial, and that the paper was not a serious piece of work.
What happens next? Will the ACM appoint a committee of the great and the good to decide whether the CACM article should be retracted? We are not talking about fraud or deception, but a bunch of silly mistakes that invalidate the claimed findings. Researchers are supposed to care about the integrity of published work, but will anybody be willing to invest the effort needed to get this paper retracted? The authors will not want to give up those 234, and counting, citations.
Update
The replication authors have been quick off the mark and posted a rebuttal of the Rebuttal.
The rebuttal of the Rebuttal has been written in the style that rebuttals are supposed to be written in, i.e., a point by point analysis of the issues raised.
Now what? I have no idea.
Replicating results using research software
The reproducibility of results, from scientific studies, has always been an important issue. Over the last few years software has become a hot topic in reproducibility circles; many researchers have an expectation that if they run the original researcher’s software, they will replicate the results. Reality has not lived up to their expectations and there has been a lot of flapping around looking for a solution. There is a solution, but first, why does the problem exist?
I have spent a lot of time porting software to different compilers (when I was in the compiler business, I wanted everybody to port their applications to the compiler I was working), different hardware (oh, the days when every major vendor had at least one distinct cpu; not like today where it’s x86, ARM, or embedded), different operating systems (umpteen flavors of Unix, all with slightly different header file contents and library behavior; the Unix wars were good for those in the porting business) and every now and again different languages (by translating).
The Wintel alliance wiped out variation in cpus and operating systems (they can still be found lurking in dark corners) and open source compilers created a near monoculture of compilers for the major languages.
The major software portability problems of 30 years ago have become rather minor. But software portability problems that once tended to be minor (at least for scientific software), have grown to become a major headache. Today’s major portability problems center around evolution of the libraries/packages being used, and longer term the evolution of the language(s) used.
Evolution has created development ecosystems where there are rampant dependencies on specific, or earlier than, or later than versions of libraries/packages. I have been out of the porting business for several decades, but talking to those doing it today, the story is the same; experience in porting from A to B is everything, second best is talking to somebody else who has gone in that direction and third best are the one-line forums such as stackoverflow.
Researchers are doing research on who-knows-what and probably have need-to-know knowledge of software and the libraries they are using, the researchers receiving a copy of the original software might know less. What is the probability that the originating and receiving researchers have exactly the same versions of libraries installed? The receiving researcher may not have any of the required libraries installed, and promptly install the latest version (which may well be more recent than the ones used by the original researcher).
A solution is available; distribute a duplicate of the researchers complete system as a container, e.g., a Docker image.
Containers solve the replication problem. But these days people want more, they actually think it should be possible to take research software and modify it to suite their own needs. Good luck with that.
Research software is written to solve a problem, often by people writing their first non-trivial programs (i.e., they are novices), with no incentive to produce something that is easy for others to use. When software is written by experienced developers, who have an incentive to build something that is easy for others to work with, multiple reimplementations are often still required to achieve something of decent quality. Creating robust software, that others can use, is very hard.
The problem with software is its invisibility; the difficulties are not visible. When the internal operations are visible, the difficulties of making changes are easier to see.
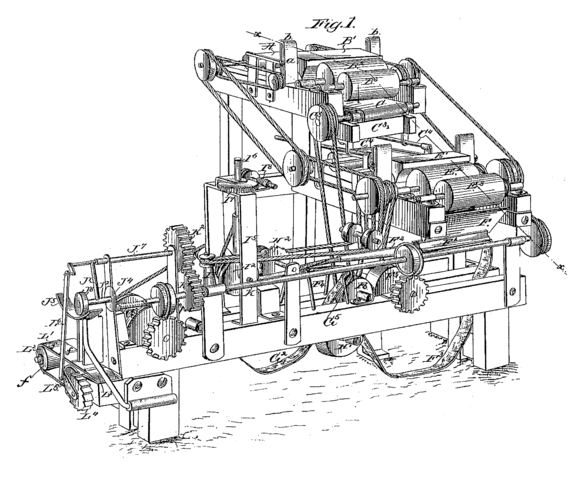
James Albert Bonsack’s cigarette rolling machine (from Wikipedia).
Replication: not always worth the effort
Replication is the means by which mistakes get corrected in science. A researcher does an experiment and gets a particular result, but unknown to them one or more unmeasured factors (or just chance) had a significant impact. Another researcher does the same experiment and fails to get the same results, and eventually many experiments later people have figured out what is going on and what the actual answer is.
In practice replication has become a low status activity, journals want to publish papers containing new results, not papers backing up or refuting the results of previously published papers. The dearth of replication has led to questions being raised about large swathes of published results. Most journals only published papers that contain positive results, i.e., something was shown to some level of statistical significance; only publishing positive results produces publication bias (there have been calls for journals that publishes negative results).
Sometimes, repeating an experiment does not seem worth the effort. One such example is: An Explicit Strategy to Scaffold Novice Program Tracing. It looks like the authors ran a proper experiment and did everything they are supposed to do; but, I think the reason that got a positive result was luck.
The experiment involved 24 subjects and these were randomly assigned to one of two groups. Looking at the results (figures 4 and 5), it appears that two of the subjects had much lower ability that the other subjects (the authors did discuss the performance of these two subjects). Both of these subjects were assigned to the control group (there is a 25% chance of this happening, but nobody knew what the situation was until the experiment was run), pulling down the average of the control, making the other (strategy) group appear to show an improvement (i.e., the teaching strategy improved student performance).
Had one, or both, low performers been assigned to the other (strategy) group, no experimental effect would have shown up in the results, significantly reducing the probability that the paper would have been accepted for publication.
Why did the authors submit the paper for publication? Well, academic performance is based on papers published (quality of journal they appear in, number of citations, etc), a positive result is reason enough to submit for publication. The researchers did what they have been incentivized to do.
I hope the authors of the paper continue with their experiments. Life is full of chance effects and the only way to get a solid result is to keep on trying.
Experiment, replicate, replicate, replicate,…
Popular science writing often talks about how one experiment proved this-or-that theory or disproved ‘existing theories’. In practice, it takes a lot more than one experiment before people are willing to accept a new theory or drop an existing theory. Many, many experiments are involved, but things need to be simplified for a popular audience and so one experiment emerges to represent the pivotal moment.
The idea of one experiment being enough to validate a theory has seeped into the world view of software engineering (and perhaps other domains as well). This thinking is evident in articles where one experiment is cited as proof for this-or-that and I am regularly asked what recommendations can be extracted from the results discussed in my empirical software book (which contains very few replications, because they rarely exist). This is a very wrong.
A statistically significant experimental result is a positive signal that the measured behavior might be real. The space of possible experiments is vast and any signal that narrows the search space is very welcome. Multiple replication, by others and with variations on the experimental conditions (to gain an understanding of limits/boundaries), are needed first to provide confidence the behavior is repeatable and then to provide data for building practical models.
Psychology is currently going through a replication crisis. The incentive structure for researchers is not to replicate and for journals not to publish replications. The Reproducibility Project is doing some amazing work.
Software engineering has had an experiment problem for decades (the problem is lack of experiments), but this is slowly starting to change. A replication problem is in the future.
Single experiments do have uses other than helping to build a case for a theory. They can be useful in ruling out proposed theories; results that are very different from those predicted can require ideas to be substantially modified or thrown away.
In the short term (i.e., at least the next five years) the benefit of experiments is in ruling out possibilities, as well as providing general pointers to the possible shape of things. Theories backed by substantial replications are many years away.

Recent Comments